Mohimanilab is moving to the Computational Medicine Department at UCLA. I am hiring graduate students and PostDocs. If you are interested in working with me, please contact me at hmohimani at mednet dot ucla dot edu.
Welcome to the Metabolomics and Metagenomics Lab
We are hiring PostDocs in the area of computational metabolomics and metagenomics

Matching Omics Big Data
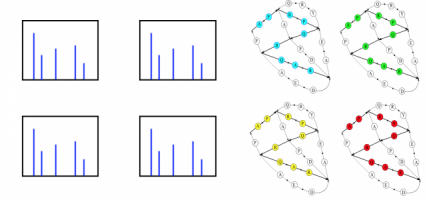
Error Tolerant Matching
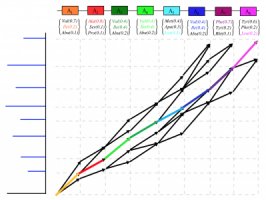
Matching Graphs
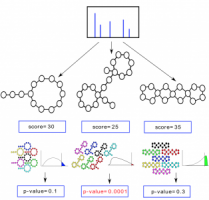
Statistical Validation
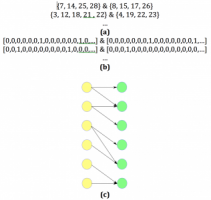
Learning Fragmentation
Hypothesis Generation

Hosein joined Carnegie Mellon University in 2017. He received his B.Sc. from Sharif University of Technology, Iran, and Ph.D. from the University of California, San Diego, working with internationally recognized computational biology leader Pavel Pevzner. He worked for two years as a Bioinformatics Scientist at Illumina before returning to UCSD as a project scientist. Hosein developed error-tolerant approaches to connect genomics and metabolomics datasets for discovering novel natural product drugs. This work resulted in the first discovery of a natural product with antibiotic activity (called Informatipeptin) in a completely automated fashion in 2013.
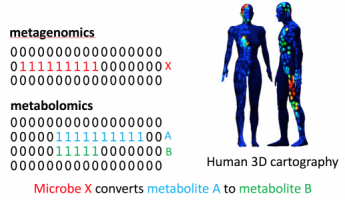
The research in MetaboloGenomics lab focuses on the development of computational metabolomics and genomics methods for natural product drug discovery. Microbial isolates from various environments produce small molecule natural products with antibiotic, antitumor and antifungal activities. Most of such molecules remain unknown. MetaboloGenomics lab is specifically interested in analyzing large scale mass spectrometry and genomics data using tools from machine learning, genome mining, signal processing, graph theory, and statistics to discover novel genetically synthesized small molecules from microbial and plant species.
News:
6/20/22 – Mihir and Mustafa’s paper “An interpretable machine learning approach to identify mechanism of action of antibiotics” has been published in Scientific Reports
01/24/22 – Dongui’s paper “TransDiscovery: Discovering Biotransformation from Human Microbiota by Integrating Metagenomic and Metabolomic Data” has been published in Metabolites journal
07/16/21 – Our lab receives an award to develop “Computational methods for nonribosomal peptide discovery” from the National Science Foundation. ($668K)
07/21/21 – Zeyuan Zuo, Liu Cao, Louis-Félix Nothia’s paper “MS2Planner: improved fragmentation spectra coverage in untargeted mass spectrometry by iterative optimized data acquisition” has been published in Bioinformatics Journal (also presented in ISMB 2021 conference)
06/17/21 – Liu Cao and Mustafa Guler‘s paper “MolDiscovery: learning mass spectrometry fragmentation of small molecules” has been published in Nature Communications
06/02/21 – Bahar Behsaz’s paper “Integrating genomics and metabolomics for scalable non-ribosomal peptide discovery” has been published in Nature Communications
04/15/21 – Mihir Mongia’s paper “Repository scale classification and decomposition of tandem mass spectral data” has been published in Scientific Reports
02/11/21 – Arash Gholamidavoodi, Sean Chang, Hyun Gon Yoo, Anubhav Baweja, Mihir Mongia and Hosein Mohimani’s paper “ForestDSH: a universal hash design for discrete probability distributions” has been published in Data Mining and Knowledge Discovery
10/25/20 – Our lab receives Computational Tool Development for Integrative Systems Biology Data Analysis award from the U.S. Department of Energy. ($1.05M)
05/20/20 – “What Is the Role for Algorithmics and Computational Biology in Responding to the COVID-19 Pandemic?”
01/28/20 – Mihir Mongia , Ben Soudry and Arash Gholamidavoodi ’s paper, “Efficient Database Search via Tensor Distribution Sensitive Bucketing” , has been accepted for PAKDD 2020.
01/09/20 – Mohsen Ferdosi and Arash Gholamidavoodi’s paper, “Measuring Mutual Information Between All Pairs of Variables in Subquadratic Complexity” , has been accepted for AISTATS 2020. Congratulations Mohsen and Arash!
10/01/19 – Our lab receives NIH New Innovator Award ($2.2M)
08/05/19 – Liu Cao and Egor Scherbin’s paper, “Association Networks”, has been accepted at mSystems. Congratulations Liu and Egor!
07/23/19 – Liu Cao’s paper, “MetaMiner“, has been accepted at Cell-Systems. Congratulations Liu! (This was also presented in RECOMB 2019 conference)
02/19/19 – Hosein Mohimani named Sloan Research Fellow
10/02/18 – Carnegie Mellon School of Computer Science: New Algorithm Efficiently Finds Antibiotic Candidates
02/60/18 – Pittsburgh Post Gazette: A new CMU algorithm can help thwart antibiotic resistance
01/30/18 – WESA, Pittsburgh’s NPR News Station: Finding A Better Antibiotic Just Got Easier, As Scientists Discover A New Way To Sift Through Data
01/22/18 – Carnegie Mellon School of Computer Sciences: Computational Method Speeds Hunt for New Antibiotics
11/18/16 – University of California San Diego: Big Data for Chemistry: New method helps identify antibiotics in mass spectrometry datasets